Data Commons
Interactive multi-omics data analysis and visualization platform
The Data Commons module allows researchers to upload, analyze, and visualize gene expression data, differential expression outputs, and dimensionality reduction results.
The platform supports standard bioinformatics file formats (.csv, .tsv, .txt) and works with both gene-level and transcript-level datasets.
Analysis Modules
The workflow consists of three integrated analysis modules:
Gene Expression Analysis
Compare expression levels across samples using interactive bar plots.
Visualize up to four genes or transcripts simultaneously, grouped by experimental conditions.
PCA Analysis
Explore sample relationships through Principal Component Analysis (PCA).
Identify clusters, detect outliers, and assess batch effects with interactive scatter plots.
Differential expression Analysis
Visualize differential expression results using volcano plots.
Adjust significance thresholds (p-value, log₂FC) in real time and export filtered gene lists for downstream analysis.
Getting Started
Step 1: Prepare Your Data Files
The platform automatically classifies uploaded files based on keywords found in the filename (case-insensitive).
File Type Detection Rules
| File Type | Required Keywords | Examples |
|---|---|---|
| Gene Expression | gene AND (count | tpm | fpkm) | salmon_gene_counts.tsvGENE_TPM_matrix.csv |
| Transcript Expression | transcript AND (count | tpm | fpkm) | transcript_counts.tsvisoform_TPM.csv |
| Differential Expression | Contains any of: diff, differential, de, expression, diffexp | differential_expression.csvDE_output.tsv |
| Sample Metadata | sample | samplesheet.csvsample_metadata.tsv |
| PCA Results | pca (or last unclassified file) | PCA_results.csvpca_coordinates.tsv |
Note: If a Differential Expression file contains the keyword
transcript, it is treated as transcript-level DE.
Step 2: Upload Files
You can analyze existing project data stored on the server or upload new files manually.
The platform supports both workflows.
Option A: Use Existing Project Data (Auto-Detected)
If your team has already uploaded datasets to the server:
- Navigate to Data Commons.
- Select your Group → Program → Project.
- The platform automatically retrieves all files associated with this project.
- Files are classified and grouped according to the detection rules
(e.g., Gene Expression, Transcript Expression, PCA, Differential Expression, Sample sheet). - Review the detected file assignments in the confirmation dialog.
- Click Continue to load the project.
This option is ideal when data for a project has been uploaded previously and follows the naming conventions defined in File Type Detection Rules.
Option B: Upload New Files Manually
If you want to analyze new datasets:
-
Click the Upload Files button.
-
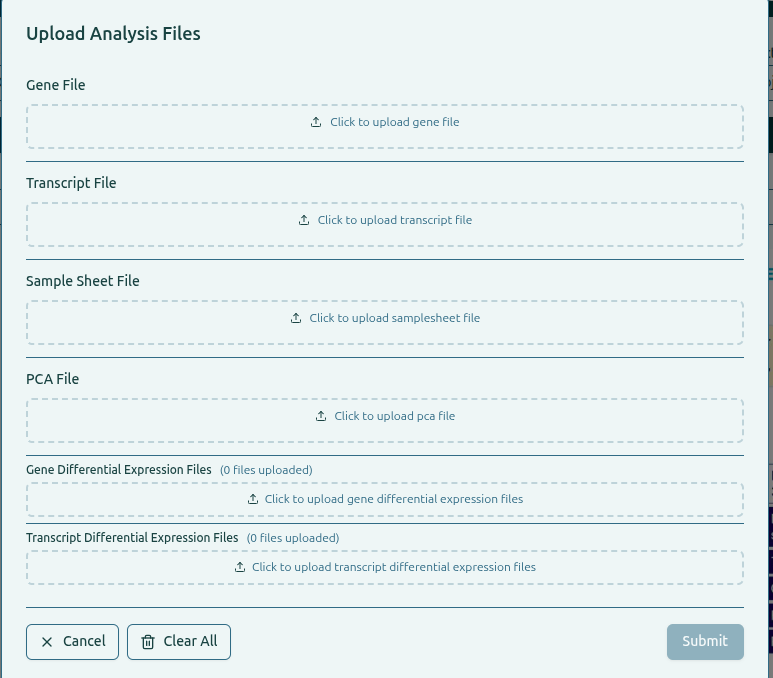
Upload files into their corresponding boxes:
- Gene File
- Transcript File
- Sample Sheet File
- PCA File
- Gene Differential Expression Files (multiple allowed)
- Transcript Differential Expression Files (multiple allowed)
-
Drag-and-drop or browse your local system to upload.
-
The platform previews each selected file and validates it.
-
Click Submit to process and index all uploaded files.
Note: All uploaded files are stored securely on the client side (browser storage) only, ensuring that your sensitive data remains private and is not exposed to any external servers during upload or preview.
Step 3: Explore Visualizations
Navigate between modules using the tab interface:
- Gene Expression: Query genes and visualize TPM, FPKM, or raw counts.
- PCA: Explore sample clustering and similarity patterns.
- Differential Expression Analysis: Adjust statistical filters in Volcano Plots to highlight significant genes.
Password-Protected Datasets
Some datasets require restricted access depending on the specific Group → Program → Project.
Password protection ensures that only authorized users can view or analyze sensitive data.
How Password Protection Works
Password security is handled entirely on the server side:
- Each project may contain a
password.txtfile inside its Group/Program/Project directory. - If this file exists, the project is considered password protected.
- The password applies only to that specific Group–Program–Project and does not affect others.
- If no
password.txtfile is present, the project loads normally without requiring authentication.
Note: The Upload Manual Files option does not include a password field, as password protection applies only to server-stored project data.
Accessing a Protected Project
When opening a project that has password.txt, a password prompt appears:
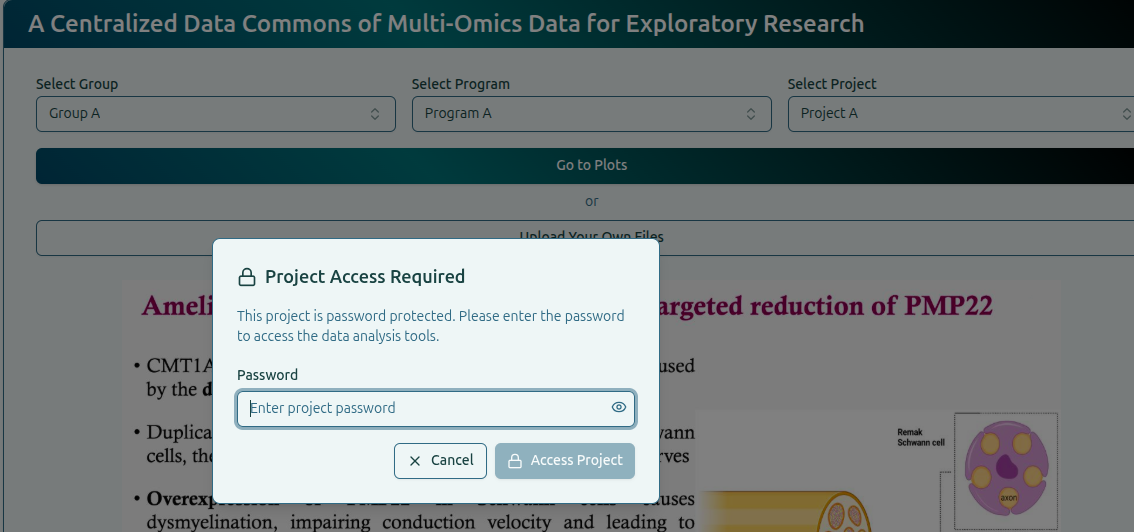
- Correct Password: The dataset is unlocked and files load normally.
- Incorrect Password: Access is denied and the project remains locked.
Security Notice: Passwords are protected, but they can be updated if the user requests.
Preview & ZIP Download (Settings Menu)
Each analysis module (Gene Expression, PCA, Volcano Plot) includes a Settings menu on the dashboard.
This menu provides two key features for reproducibility and data sharing.
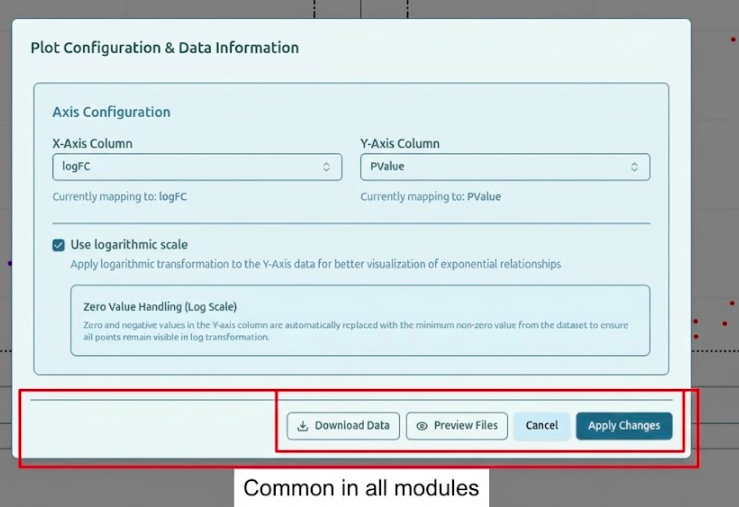
1. Preview Files
View all files associated with the current analysis:
- Raw uploaded files
- Processed intermediate tables
- Statistical result files (e.g., DE tables)
- Plot-specific data matrices
This preview helps verify data integrity and confirm which files were used in the analysis.
2. Download All Files as ZIP
Click Download ZIP inside the Settings menu to export a complete package containing:
- Processed analysis tables (CSV)
- Expression matrices
- Sample sheet files
File Requirements Checklist
Before uploading, ensure:
- Format: Files end with
.csv,.tsv, or.txt. - Naming: Required filename keywords are included (see Step 1).
- Structure: Single header row; no merged cells or comments.
- Data: Numeric fields contain valid numbers (no
NA,Inf, or text).